The Melbourne Advanced Gene Editing Centre (MAGEC) was established in 2014 as one of the nodes of Phenomics Australia. Located at the Walter and Eliza Hall Institute of Medical Research in Melbourne, their aim is to develop cutting-edge gene editing technologies and to use these methods to generate gene-edited mouse models for the Australian research community. Researchers at the centre have adapted and refined the CRISPR/Cas9 system within their gene-editing pipeline to successfully generate more than 220 unique mouse models for research and pre-clinical models of human disease.
~ Rhyannah Hamer
The CRISPR/Cas9 system was first discovered in bacterial and archaeal genomes as a form of acquired immunity towards foreign genetic sequences introduced by invading pathogens. The key components of the CRISPR/Cas9 system feature a single guide RNA (sgRNA) and a CRISPR associated endonuclease such as Cas9, derived from the bacterial strain Streptococcus pyogenes. Cas9 endonuclease activity is directed by a unique 20 nucleotide sequence present on sgRNAs towards almost any genomic sequence, resulting in a double-stranded DNA break. When the CRISPR/Cas9 system is adapted to mammalian cells, it becomes possible to utilise the formation of these specific double-stranded breaks to generate gene-edited cells and animal models. These can be used for basic research and pre-clinical models of human diseases.
‘The use of the CRISPR/Cas9 system in the zygote has revolutionised speed, simplicity and efficiency in producing genetically modified mice.’

It is well accepted that simple modifications such as gene deletions and small insertions are highly reproducible. However, the use of this methodology to develop mouse models with more complex genetic modifications, such as floxed or large knock-in alleles, has been associated with much lower success rates.
The key reason for the low efficiency of CRISPR/Cas9-mediated generation of floxed alleles is due to the methodology outlined by the initial landmark studies describing the use of CRISPR/Cas9 in mouse zygotes1, 2. These studies described the use of two sgRNAs and two loxP-encoding single-stranded oligonucleotide donors (ssODN) to generate floxed alleles. This approach entails the following limitations:
- High Cas9 cleavage activity combined with the use of two sgRNAs often results in the deletion of the entire genomic region flanked by the sgRNAs (Figure a (i))
- Homology-directed repair (HDR)-mediated incorporation of loxP-encoding ssODNs can occur on different alleles, leading to unusable single loxP integration sites and potential false positives (Figure a (ii))
- The palindromic nature of loxP sequences often contributes to poor ssODN synthesis and frequent sequence errors, especially when long oligonucleotide donors are synthesised.
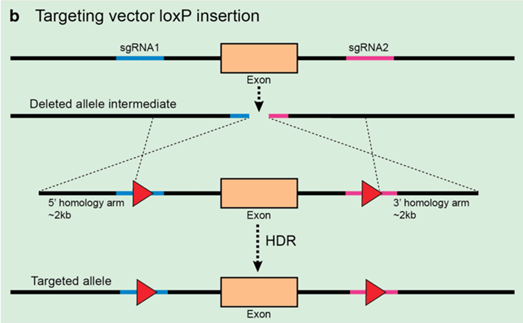
During extensive optimisations in the MAGEC laboratory, researchers discovered they could avoid these limitations by using large targeting vectors with extended homology arms as DNA donors, rather than ssODN donors, to facilitate high efficiency incorporation of loxP sites with high fidelity3. Specifically, this involved utilising large targeting vectors featuring 2 kb or more of homology arms and two sgRNA recognition sites (Figure b) to generate more than 25 unique floxed alleles across multiple genetic loci.
With this approach, they were able to exploit the high Cas9 cleavage activity to delete genomic sequences in between the two sgRNA recognition sites. This facilitated the replacement of the deleted sequence with a loxP-flanked allele via HDR-mediated incorporation of large targeting vectors in a single step.

In their first year of operation in 2014, the majority of project completions in the MAGEC laboratory were simple gene deletion mouse models, with relatively few of the more complex targeting models being made. However, rapid optimisation of DNA donors and Cas9 gene-editing reagents led to a surge in gene-targeting efficiency and the generation of highly complex mouse models, consisting of floxing (represented in green in Figure c) and other large gene targeting insertions and replacements (represented in orange in Figure c). These technological advancements have caused a large demand in generating new complex models for academic research laboratories and industry partners, both within Australia and internationally.
Approximately 50% of project completions in the past two years comprised of complex mouse models, with a total of 78 such models completed to date. The generation of such complex mouse models has become vital for advancing research in the Australian research community. Through the assistance of Phenomics Australia, nodes such as the MAGEC at WEHI are able to provide Australian researchers with modern and high quality technology that consequently helps to better the health of Australians.
For over 10 years, Phenomics Australia has developed into a strong joint venture that has provided hundreds of researchers with the means to carry out vital biomedical research. The network continues to involve itself with major development set by the government to consistently provide researchers with cutting-edge technology.
Recently, the Medical Research Future Fund has been established for the support of health and medical research, investing a total of $20 billion. With support from such emerging developments, Phenomics Australia plans on continuing providing the highest quality infrastructure to fulfil their vision of enabling genomic knowledge and technology to allow Australians to live longer and better.
Figures:
Figure a
Kueh AJ et al. Cell Death Differ. 2017; 24(10): 1821-1822
Schematic of loxP insertions using the ssODN method.
(a) The ssODN loxP insertion method utilizes Cas9-mediated double-stranded breaks at two sgRNAs positions flanking the targeted exon to stimulate the insertion of two loxP-encoding ssODNs in two independent HDR events. This method is prone to (i) the formation of a deleted allele, (ii) single loxP insertions and (iii) error-prone loxP insertions.
Figure b
Kueh AJ et al. Cell Death Differ. 2017; 24(10): 1821-1822
Schematic of loxP insertions using the large targeting vector method.
The targeting vector loxP insertion method utilizes Cas9 double-stranded breaks at the two sgRNA positions to excise intervening DNA sequence, resulting in the formation of a deleted allele intermediate. Before the deleted allele intermediate is repaired by NHEJ, the targeting vector with appropriate homology arms can introduce a floxed allele in a single HDR event.
Figure c
Pie charts depicting the percentages of project completions according to project type from 2014 to 2018 in the MAGEC laboratory.
References:
1. Wang H et al. Cell 2013; 153: 910–918.
2. Yang H et al. Cell 2013; 154: 1370–1379.
3. Kueh AJ et al. Cell Death Differ. 2017; 24(10): 1821-1822